Hyper-parameter tuning
xgboost has quite some number of parameters to tune. It is a daunting task to perform global optimisation in such a large parameter space. A couple of optimisation approaches are tried for the tuning of hyper-parameters.
- Randomised search on hyper-parameters
- Bayesian optimisation over hyper-parameters.
- Evolutionary algorithm on hyper-parameters tuning.
Parameter space for Randomised search/Evolutionary algorithm
n_estimators: [100, 200, 500, 1000, 2000]
learning_rate: [0.01, 0.02, 0.05, 0.1, 0.2]
gamma: [0.0, 0.01, 0.02, 0.05]
min_child_weight: [1, 3, 5]
max_depth: [3,5,7,9]
subsample: [0.4, 0.6, 0.8, 1.0]
colsample_bytree: [0.4, 0.6, 0.8, 1.0]
Parameter space for Bayesian optimisation
n_estimators: (100, 2000, 'log-uniform')
learning_rate: [1.e-2, 2.e-1, 'log-uniform']
gamma: [0.0, 0.05, 'uniform']
min_child_weight: (1, 5, 'uniform')
max_depth: (3, 9, 'uniform')
subsample: [0.4, 1.0, 'uniform']
colsample_bytree: [0.4, 1.0, 'uniform']
For randomised and Bayesian search, 200 points in the parameter space are used for optimisation. For evolutionary algorithm, population size and number of generations are set to 50 and 5, respectively, resulting 179 points in the parameter space.
The following table summarises the results.
| default | Random | Bayes | Evolutionary | |
|---|---|---|---|---|
| MSE | 610.3 | 572.6 | 694.8 | 480.1 |
| R2 | 0.8240 | 0.8349 | 0.7996 | 0.8615 |
| score | inf | 1.9e148 | 2.3e101 | 2.9e125 |
The following plots show true RUL versus predicted RUL and true RUL versus residuals (= true - predicted) on validation and test datasets with the hyper-parameter set obtained by Evolutionary algorithm.
| validation data | test data |
|---|---|
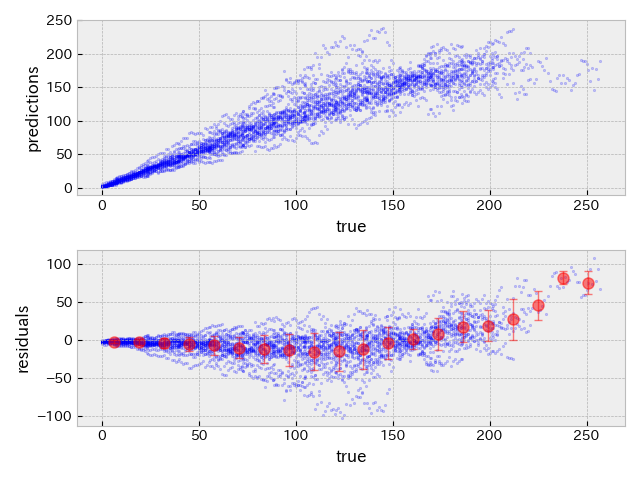 |
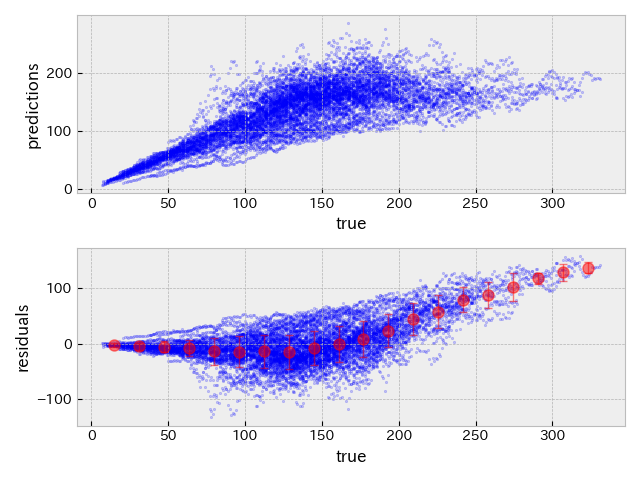 |
